Chapter 9 Logistic Regression
For estimating probability of a qualitative variable. See MLLN Notes for exploratory plots and motivation.
9.1 Simple Logistic Regression
We use the Default data from library("ISLR2") package. We also load library(scales) package for transparency with colours.
##
## Attaching package: 'scales'## The following object is masked from 'package:purrr':
##
## discard## The following object is masked from 'package:readr':
##
## col_factor##
## Attaching package: 'ISLR2'## The following objects are masked _by_ '.GlobalEnv':
##
## Boston, Hitters## The following object is masked from 'package:MASS':
##
## Boston## The following objects are masked from 'package:ISLR':
##
## Auto, CreditWe use the generalised linear model function glm() with family = "binomial".
data(Default)
glm.fit = glm( as.numeric(Default$default=="Yes") ~ balance, data = Default, family = "binomial")
summary(glm.fit)##
## Call:
## glm(formula = as.numeric(Default$default == "Yes") ~ balance,
## family = "binomial", data = Default)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -2.2697 -0.1465 -0.0589 -0.0221 3.7589
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -1.065e+01 3.612e-01 -29.49 <2e-16 ***
## balance 5.499e-03 2.204e-04 24.95 <2e-16 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 2920.6 on 9999 degrees of freedom
## Residual deviance: 1596.5 on 9998 degrees of freedom
## AIC: 1600.5
##
## Number of Fisher Scoring iterations: 8summary(glm.fit$fitted.values)## Min. 1st Qu. Median Mean 3rd Qu. Max.
## 0.0000237 0.0003346 0.0021888 0.0333000 0.0142324 0.9810081Plot
#Raw Data
plot(Default$balance, as.numeric(Default$default=="Yes"),col="red",xlab="balance",ylab="default")
#Fitted Values
points(glm.fit$data$balance,glm.fit$fitted.values, col = "black", pch = 4)
#Model Curve
curve(predict(glm.fit,data.frame(balance = x),type="resp"),col="blue",lwd=2,add=TRUE)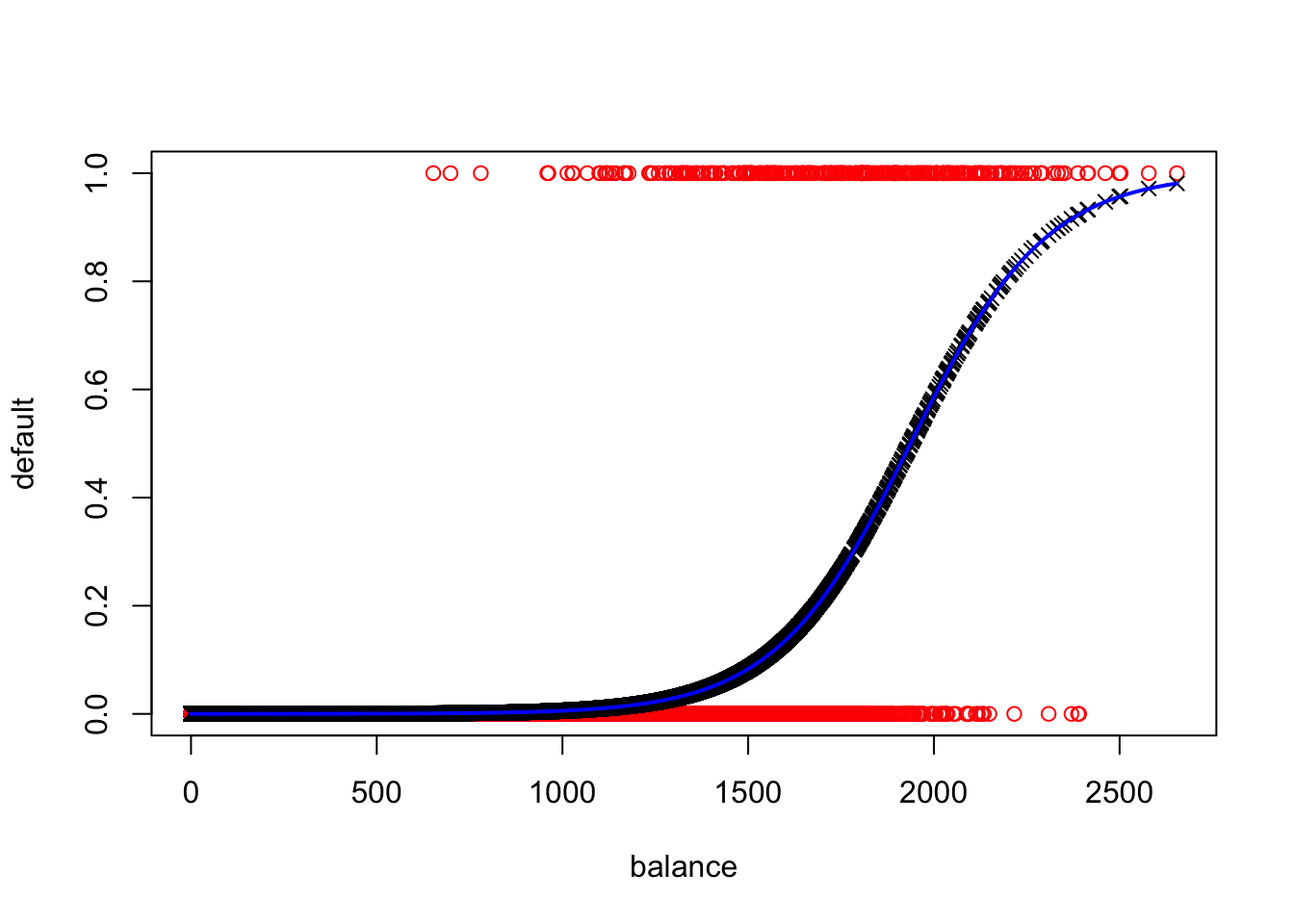
Predicting \(\hat{p}(X)\)
X = 1300
p.hat = exp(sum(glm.fit$coefficients*c(1,X)))/(1+exp(sum(glm.fit$coefficients*c(1,X))))
p.hat## [1] 0.029234419.2 Multiple Logistic Regression
Pairs Plot (Factor Variable Coloured)
pairs(admit[,2:4], col=admit[,1]+2, pch=16)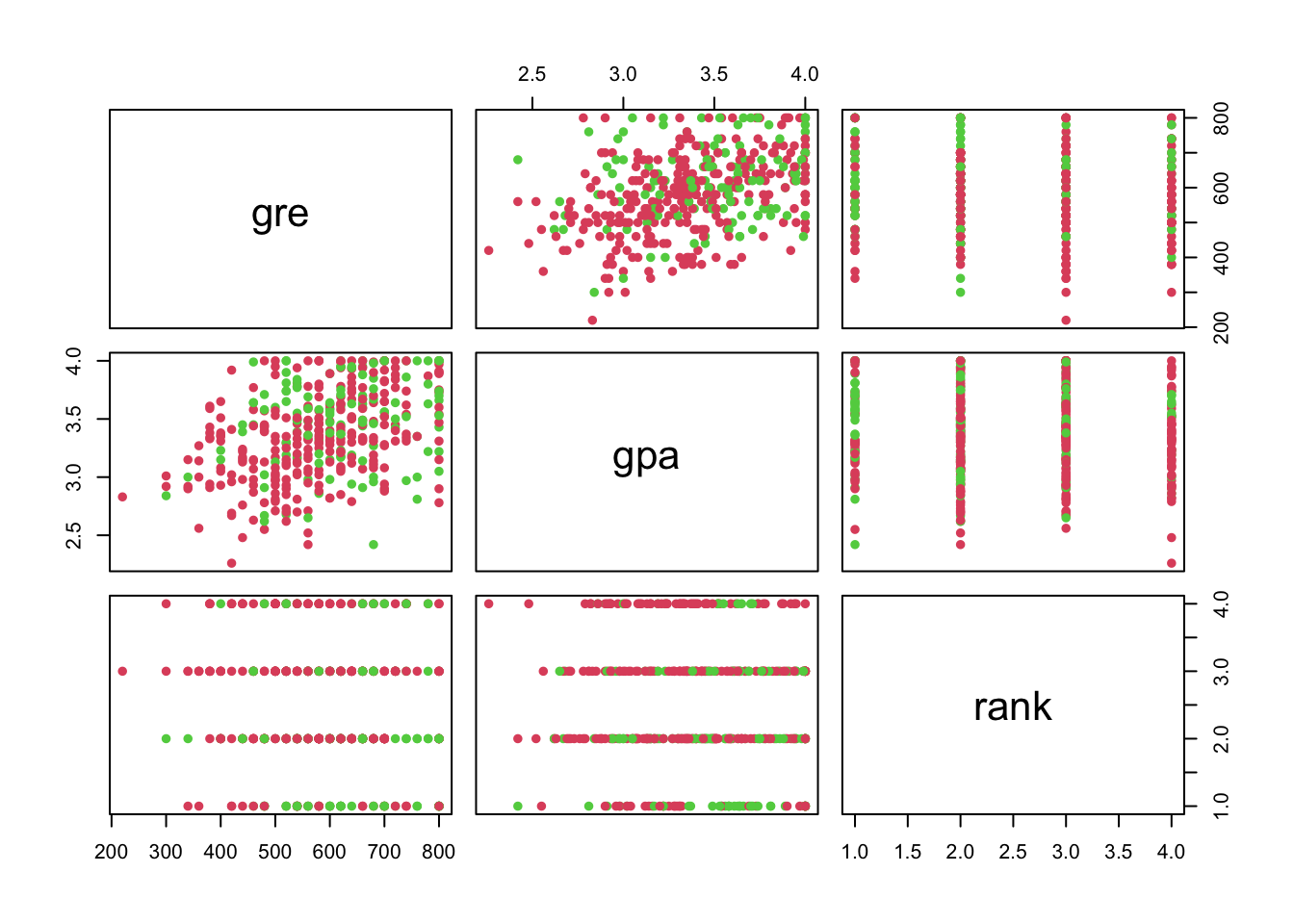
Making a Variable a Factor
admit$rank = factor(admit$rank)Practical 4 Example
glm.fit = glm( admit ~ ., data = admit, family = "binomial")
summary(glm.fit)##
## Call:
## glm(formula = admit ~ ., family = "binomial", data = admit)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -1.6268 -0.8662 -0.6388 1.1490 2.0790
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -3.989979 1.139951 -3.500 0.000465 ***
## gre 0.002264 0.001094 2.070 0.038465 *
## gpa 0.804038 0.331819 2.423 0.015388 *
## rank2 -0.675443 0.316490 -2.134 0.032829 *
## rank3 -1.340204 0.345306 -3.881 0.000104 ***
## rank4 -1.551464 0.417832 -3.713 0.000205 ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for binomial family taken to be 1)
##
## Null deviance: 499.98 on 399 degrees of freedom
## Residual deviance: 458.52 on 394 degrees of freedom
## AIC: 470.52
##
## Number of Fisher Scoring iterations: 49.3 Inference
Calculating Probability
Use predict() function with type = "response" or type = "resp". The type = "response" option tells R to output probabilities of the form P(Y=1|X), as opposed to other information such as the logit. Do not include any data to calculate predictions for training data (used to plot the curve).
glm.probs = predict(glm.fit, type = "response")Single Prediction/Multiple New Prediction
#Data For Prediction
testdata = data.frame(gre = 380, gpa = 3.61, rank = 3)
testdata$rank = factor(testdata$rank)
#Single Prediction
predict(glm.fit, testdata, type="resp")## 1
## 0.1726265Predictions
Round probability to get a prediction.
#Prediction
glm.pred = round(glm.probs)
#Confusion Table
table(glm.pred, admit$admit)##
## glm.pred 0 1
## 0 254 97
## 1 19 30#Ratio of correct predictions on test data
mean(glm.pred == admit$admit)## [1] 0.71